How to Determine Which Restriction Enzyme to Use
B for BamHI enzyme E for EcoRI enzyme H for HindIII enzyme and L- for Lambda DNA uncut. Remember to substitute dH2O for enzyme in the uncut controls.
335 Recdna Restriction Enzymes
Lambda or adenovirus-2 DNA with restriction enzyme to test enzyme viability.
. However some produce blunt ends. Calculate the volume of enzyme you will need to add if you use 1 unit per microgram of DNA or 5 units total For example if the enzyme concentration is 1 Unitul and I need 5 Units then I would need. DNA genomic is digested by restriction enzyme then an oligo-targeter is added for detection.
Up to 24 cash back In this experiment you will determine the relative locations of three restriction enzyme cleavage sites on a circular plasmid DNA. Enzyme 1 cleaves the plasmid once. Set up each reaction with all reagents except the restriction enzyme in each.
Restriction digests are commonly used to confirm the presence of an. By clicking the scissors icon on the right-hand navigation bar access Digests. The required volumes of enzyme and buffer in each reaction tube are given below.
Use a new tip for each reagent. Each enzyme recognizes one or a few target sequences and cuts DNA at or near those sequences. If the control DNA is cleaved and the experimental DNA resists cleavage the two DNAs can be mixed to determine if an inhibitor is present in the experimental sample.
Set up the reaction using the following scheme. The majority of restriction enzymes cut 6 base pair palindromes as seen with the examples above. 1 Determine the amount total ug and total ul of DNA to be digested.
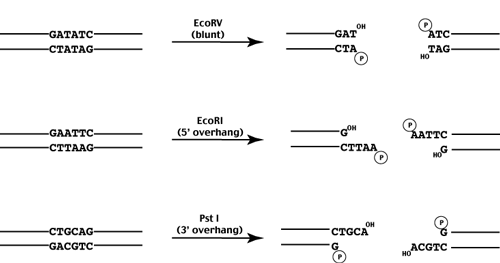
DNA ligase is a DNA-joining enzyme. Many restriction enzymes make staggered cuts producing ends with single-stranded DNA overhangs. There are many different ways to use the tool.
Diagnostic restriction digests are comprised of 2 separate steps. Determine an appropriate reaction buffer by reading the instructions for your enzyme. These restriction enzymes are able to scan along a length of DNA looking for a particular sequence of bases that they recognize.
These enzymes which are usually found in bacteria and other prokaryotes are considered as one of the most important tools in recombinant DNA technology since they can. DNA restriction buffer water and. Restriction enzymes are DNA-cutting enzymes.
An alternative approach is to digest with two different enzymes in three stages. Restriction enzymes use an enzyme originating from a bacterium that has the capability of recognizing particular base sequences in the DNA at that site. Label four 15ml tubes in which you will perform restriction digestion.
When selecting restriction enzymes for use in a cloning experiment it is important to. Each team should set up five digestions in the order given above. The plasmid has been cleaved with two restric- tion enzymes.
The enzyme volume must be 10 or less. 4 Choose a total volume for the reaction. Enzyme 2 cuts the plasmid twice.
Determine which enzymes will produce ends compatible with the selected vector confirm that recognition sites do not occur within the DNA fragment to be cloned and examine the methylation sensitivity of your selected enzyme to confirm that host methylation by dam andor dcm methylases will. Select restriction enzymes to digest your plasmid. Pro-Tip To determine which restriction enzymes will cut your DNA sequence and where they will cut use a sequence analysis program such as Addgenes Sequence Analyzer.
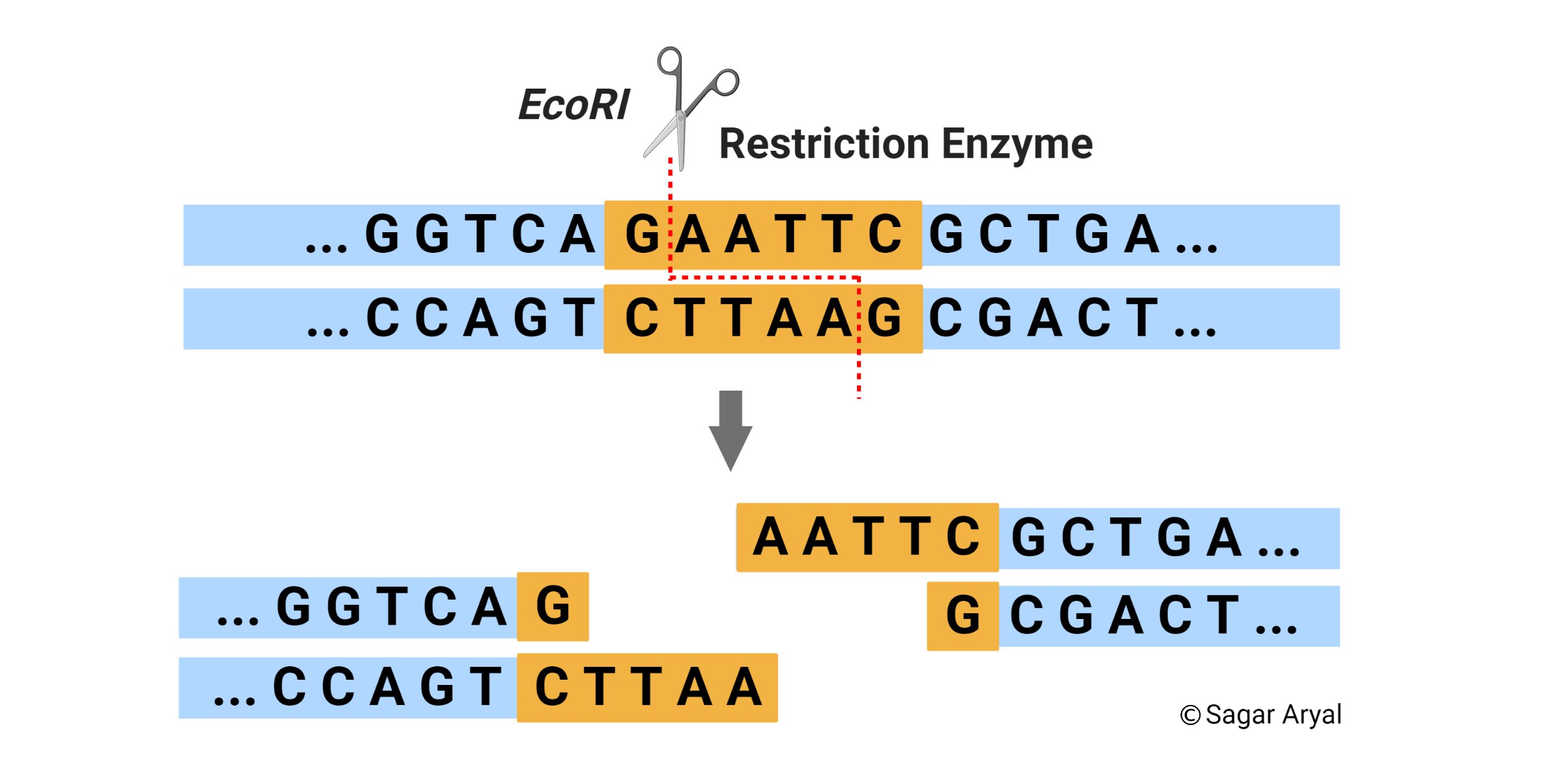
Assume that the Enzyme 1 site is at position O. Search by the number of cuts 1 find and left-click on AseI to select this enzyme and hit Run Digest. A restriction enzyme or restriction endonuclease is an enzyme that cleaves DNA into fragments at or near specific recognition sites within the molecule known.
In the examples above EcoRI produces ends with single-stranded 5 overhangs and PstI produces ends with 3 overhangs. Identify enzymes by the cut sites or the name of their named or number of them. A restriction enzyme restriction endonuclease is a special enzyme that recognizes a specific sequence of nucleotides and cleaves DNA at that specific site restriction site or target sequence.
5 Units1 Unitul 5 ul My sample needs. Then get the Nalgene ice tray with the enzymes and add the enzymes to each reaction. First enter your DNA sequence into NEBcutter.
Most restriction enzymes cut their corresponding restriction sites in a staggered fashion leaving single-stranded overhangs. Special enzymes termed restriction enzymes have been discovered in many different bacteria and other single-celled organisms. 3 Determine how many ul of enzyme to use using the enzyme concentration.
How To Find Restriction Sites Using Gene Construction Kit. Open and switch the view to Linear Map. One unit of restriction endonuclease activity is defined as the amount of enzyme required to produce a complete digest of 1 µg of substrate DNA or fragments in a.
DNA genomic is used for PCR amplicon. The word restriction in restriction enzymes refers to the restriction or limiting of the DNA from any foreign DNA at different restriction sites site in the DNA where a particular. How do you know how much restriction enzymes to use.
Restriction enzymes 05 l each Total volume 200 l 4. Using table below add reagents to each tube in this order. Open the Digests button navigate to New Digest and specify NEB and Double Cutters in the settings.
First with restriction enzyme A Second with restriction enzyme B Third with both enzymes A B. 2 Use the ug amount of DNA to determine how many enzyme units to use. This recognition site or sequence is generally from 4 to 6 base pairs in length.
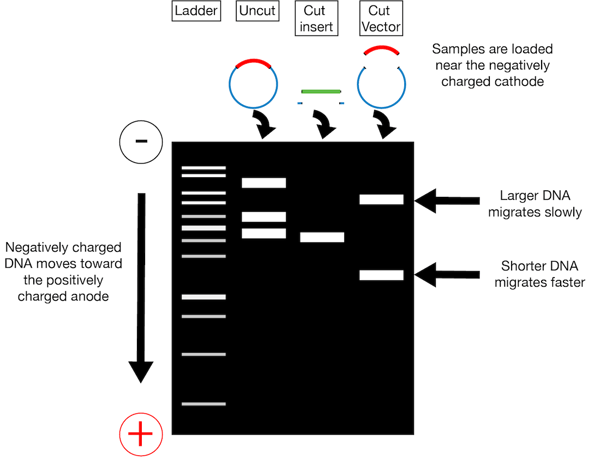
NEBcutter is a free tool that is used for restriction enzyme analysis. Also you will need to calculate the volume of dH2O required to bring each reaction to the total volume indicated 20 ul or 50 ul depending on the enzyme. 1 incubating your DNA with restriction enzymes which cleave the DNA molecules at specific sites and 2 running the reaction on an agarose gel to determine the relative sizes of the resulting DNA fragments.
The inputs are three sets of. Control DNA DNA with multiple known sites for the enzyme eg. In this video you will learn how to find out which restriction enzymes can be used to cut your DNA sequence a specific number of times.

Restriction Enzyme Analysis How To Make The Cut

How To Recognize A Recognition Site For A Restriction Enzyme Youtube
Restriction Enzyme Restriction Endonuclease
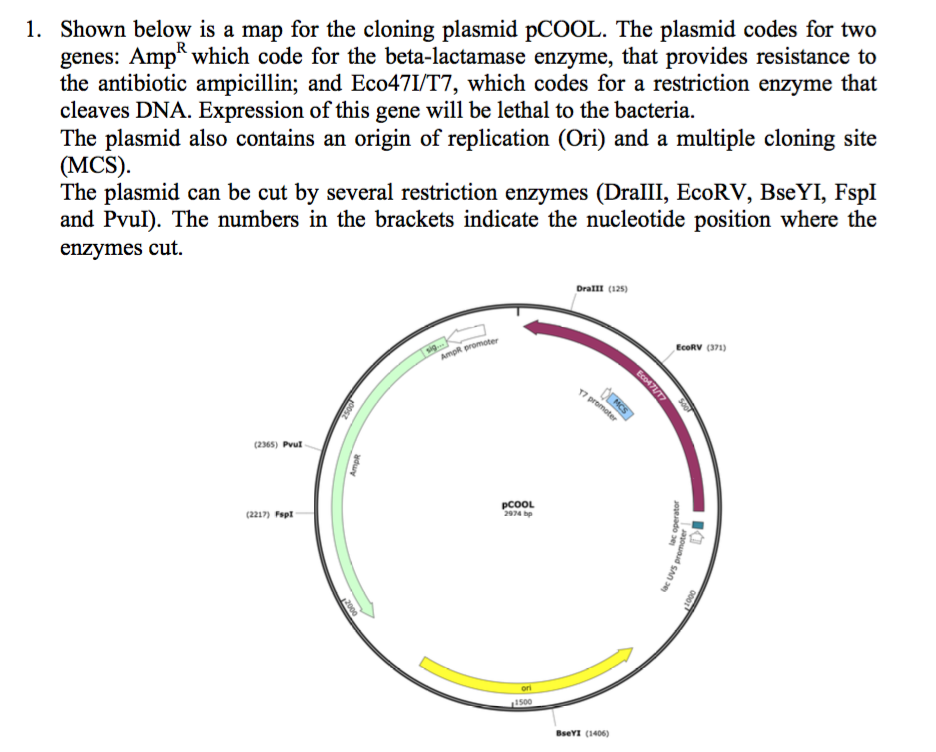
Solved 1 Shown Below Is A Map For The Cloning Plasmid Chegg Com
Bio 2960 Lab Restriction Digest Computer Lab

Plasmids 101 Restriction Cloning

Restriction Enzyme An Overview Sciencedirect Topics
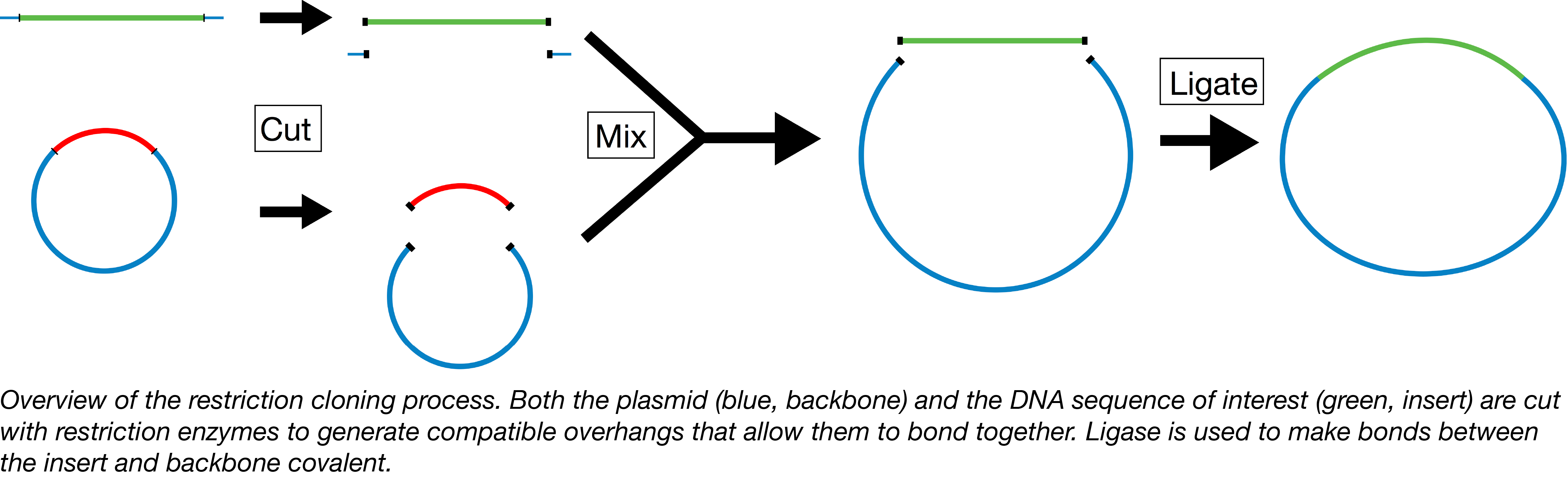
Plasmids 101 Restriction Cloning

Ap Biology Restriction Enzyme Digests On Circular Plasmids Youtube
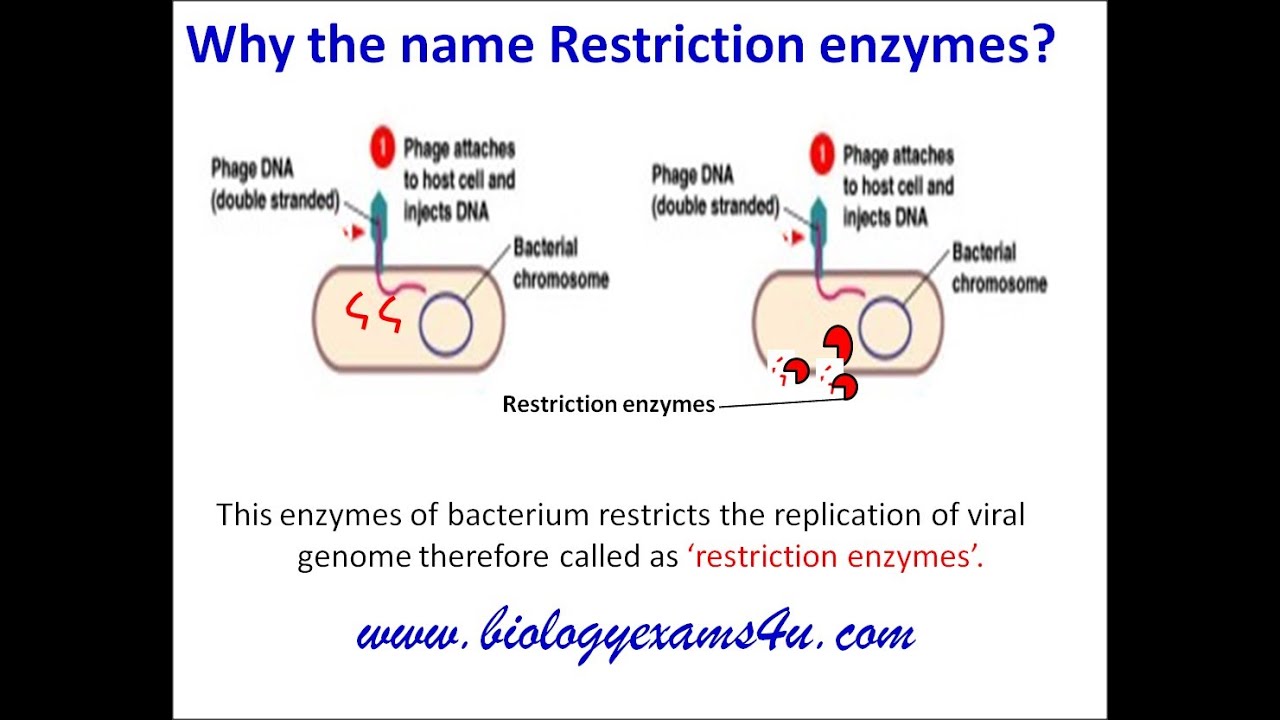
Restriction Enzymes Definition Types And Cut Patterns Youtube
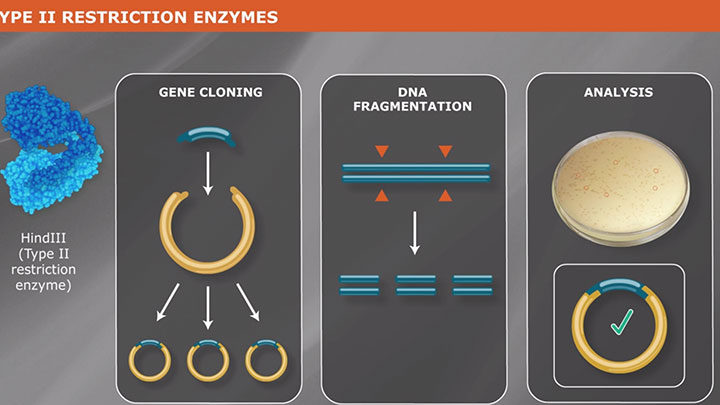
Restriction Enzyme Digestion Neb

Choosing Proper Restriction Enzymes Based On Defined Criteria For Pcr Download Scientific Diagram
Restriction Enzymes How Is Dna Manipulated

Activity 3 Restriction Enzyme Analysis

How To Recognize A Recognition Site For A Restriction Enzyme Youtube

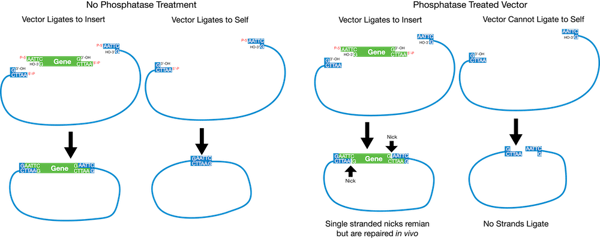

Comments
Post a Comment